
Identifying novel therapeutic targets related to TDP-43 loss : Velina Kozareva
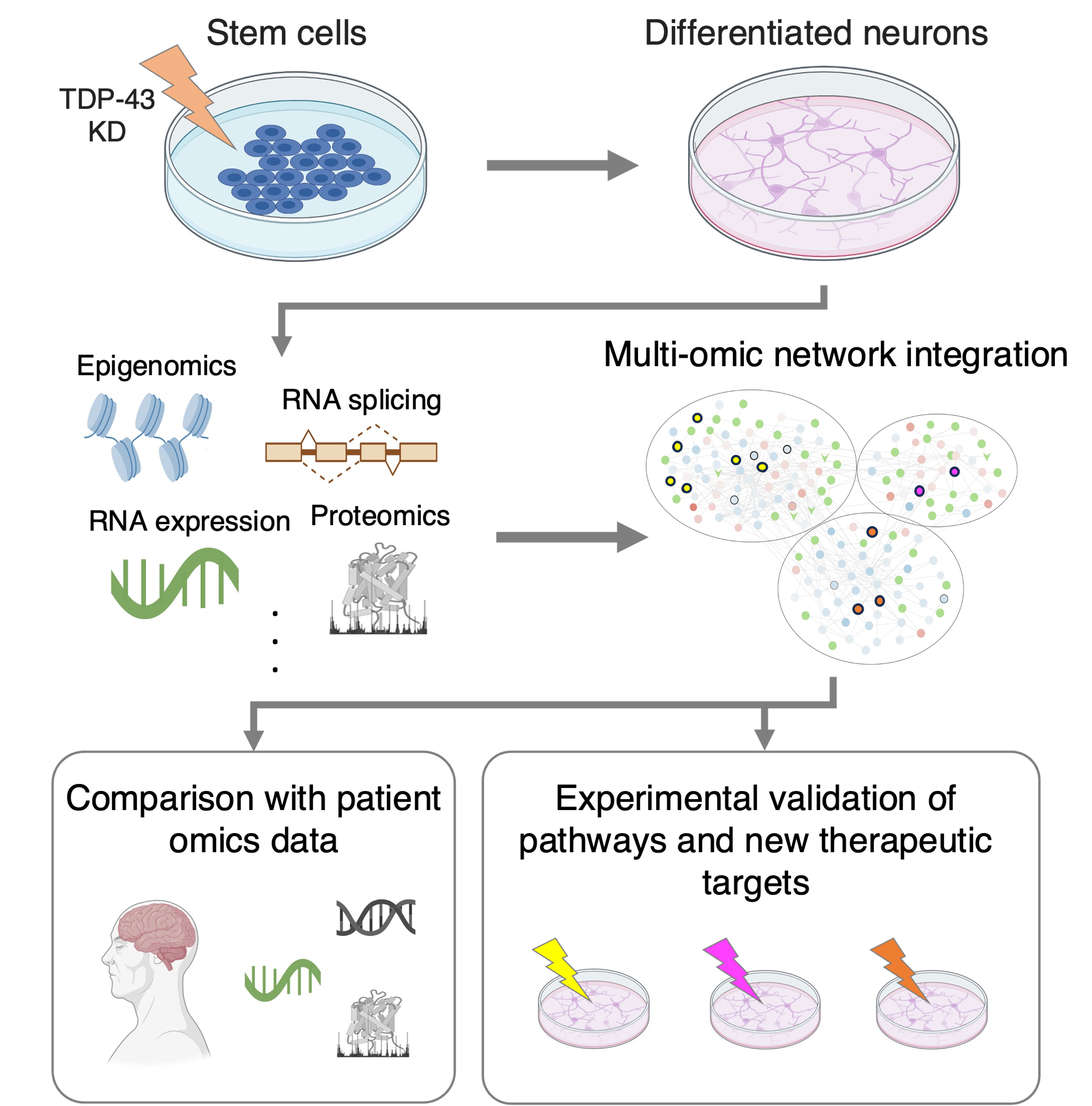
Despite the diversity of ALS, almost all patients have one thing in common: the protein TDP-43 stops performing its normal functions in the nucleus, leading to disrupted splicing and decreased expression of many genes. Several of the most prominent genes affected by this loss, such as STMN2 and UNC13A, are already being investigated as potential therapeutic targets. However, we still lack a complete understanding of all the processes impacted downstream of TDP-43 loss – and how these pathway defects lead to motor neuron death. Identifying and characterizing these pathways is crucial for developing new treatments that potentially target multiple aspects of neuronal degeneration in ALS. In collaboration with the groups of Sarah Kargbo-Hill at U. Michigan and Michael Ward at the NIH, we are applying computational models to better understand these disrupted pathways, using data from stem-cell derived neurons that were altered to reduce nuclear levels of TDP-43. Our analysis combines multiple data types – including gene expression and splicing changes, along with protein abundance levels – into integrated networks that can help characterize pathway-level effects. So far, our models have revealed significant links between TDP-43 splice targets and global protein expression changes, highlighting defects in processes related to neuronal activity and regeneration, protein degradation, and cellular metabolism. Importantly, we’ve found similar pathway changes in data generated from post-mortem patient neurons, and we are working to identify associations between these processes and known genomic risk factors for disease. With our colleagues, we are also currently investigating how changing the expression of specific genes involved in these pathways modifies the growth and function of stem-cell derived neurons. In the future, we plan to use this experimental platform to test ways to restore normal functions in these cells, with the ultimate goal of discovering novel mechanisms and gene combinations that can be therapeutically targeted to treat ALS.
Velina Kozareva is a PhD Student in the Computational and Systems Biology program at MIT, advised by Professor Ernest Fraenkel. Her research focuses on developing and applying computational models for integration of multiple data types, with a particular interest in understanding neurodegenerative diseases. She has previously worked to create methods for analyzing high-throughput genomics data at both the Broad Institute of MIT and Harvard and in industry.



